Research

Dual-inhibition of DNA Gyrase and Topoisomerase IV
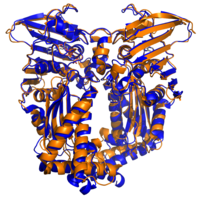 DNA Gyrase and Topoisomerase IV are bacterial type II topoisomerase enzymes responsible for manipulating the degree of supercoiling in DNA. These enzymes are established antibiotic targets; the flouroqiunolone antibiotics act by binding to the DNA binding region of the enzymes.
DNA Gyrase and Topoisomerase IV are bacterial type II topoisomerase enzymes responsible for manipulating the degree of supercoiling in DNA. These enzymes are established antibiotic targets; the flouroqiunolone antibiotics act by binding to the DNA binding region of the enzymes.
In this project we are attempting to identify potent inhibitors of the ATP binding site of these enzymes. The ATP binding site is located in the GyrB domain of DNA gyrase and the ParE domain of topoisomerase IV. Dual-inhibitors of these two targets would be good candidates as antibiotics as they will have a novel mode of action to any currently available antibiotics.
Identification of tool compounds to probe insulin receptor: insulin-like growth factor 1 receptor (IR:IGF-1R) hybrid dimer formation
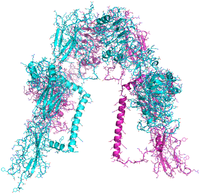 Type 2 diabetes is an increasingly common disorder. Around 2.9 million people in the UK are affected by diabetes and there are estimates of another 850 000 undiagnosed cases. Insulin resistance, which precedes the development of type 2 diabetes by many years, is also an independent risk factor for the development of premature cardiovascular disease.
Type 2 diabetes is an increasingly common disorder. Around 2.9 million people in the UK are affected by diabetes and there are estimates of another 850 000 undiagnosed cases. Insulin resistance, which precedes the development of type 2 diabetes by many years, is also an independent risk factor for the development of premature cardiovascular disease.
It is well established that IGF-1R and IR are able to form hybrid receptors consisting of an IR αβ heterodimer and an IGF-1R αβ heterodimer. IR:IGF-1R stoichiometry is a critical determinant in endothelial cell insulin sensitivity, nitric oxide bioavailability and vascular repair. IGF-1R has a negative effect on insulin signalling due to formation of insulin resistant hybrid receptors with IR.
Work in the group has developed a homology model of the IR:IGF-1R hybrid structure and used hotspot prediction software to predict the key regions involved in hybrid dimer formation. Utilising the eHiTS and AUTODOCK virtual high-throughput screening tools we have identified a number of small molecules which are predicted to inhibit formation of the IR:IGF-1R hybrids. We have developed a Bioluminescence Resonance Energy Transfer assay to measure the interaction of the IR and IGF-1R monomers and thereby quantify the action of our small molecule probes on inhibiting dimer formation.
Synthesis and Biological Evaluation of Novel KNa1.1 Channel Inhibitors to Treat KCNT1-Related Epilepsy
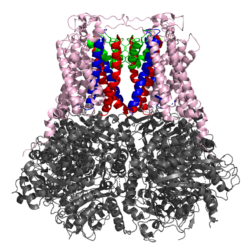 The KCNT1 gene encodes for a sodium-gated potassium channel known as KNa1.1 (Slack). Gain-of-function missense mutations of KCNT1 give rise to the rare neurological conditions, epilepsy of infancy with migrating focal seizures and autosomal dominant frontal lobe epilepsy. KCNT1-related epilepsies often respond poorly to frontline antiepileptics. The pore-blocker quinidine has shown varying efficacy clinically, due to its non-selectivity and significant risk of cardiac arrythmia.
The KCNT1 gene encodes for a sodium-gated potassium channel known as KNa1.1 (Slack). Gain-of-function missense mutations of KCNT1 give rise to the rare neurological conditions, epilepsy of infancy with migrating focal seizures and autosomal dominant frontal lobe epilepsy. KCNT1-related epilepsies often respond poorly to frontline antiepileptics. The pore-blocker quinidine has shown varying efficacy clinically, due to its non-selectivity and significant risk of cardiac arrythmia.
The aim of this project is to develop potent Slack channel inhibitors, which overcome the issue of selectivity. We have identified a wide variety of hit-matter, through computational high-throughput screening and cell-based assays. We are now exploring the effect that functional changes have on the in vitro potency and selectivity of these inhibitors.
